MedTagger is a collaborative framework for annotating medical datasets.
Main goal of this project was to design and develop software environment, which helps in aggregation and labeling huge datasets of medical scans, powered by idea of crowdsourcing. Platform also provides mechanism for label validation, thus making produced datasets of labels more reliable for the future use.
MedTagger is still under heavy development, so please keep in mind that many things may change or new versions may not be fully backward compatible. Contact with us directly in case you want to use our work :)
Documentation for the MedTagger can be found here.
MedTagger consists of two main parts:
frontend - User Interface application written in TypeScript & Angular (more),backend - system's architecture and API written in Python (more).To set up MedTagger locally you can use Vagrant virtual machine:
$ vagrant upThen follow up with our documentation. Default development account is:
admin@medtagger,medtagger.MedTagger can be set up easily with Docker-Compose:
$ docker-compose upMore about setting up environment with Docker-Compose can be found here.
Together with Docker-Compose setup, you can use Jupyter Notebook server to easily analyse collected annotations. Here you can find some examples that shows how to do so.
More information about setting up local Jupyter Notebook session can be found here.
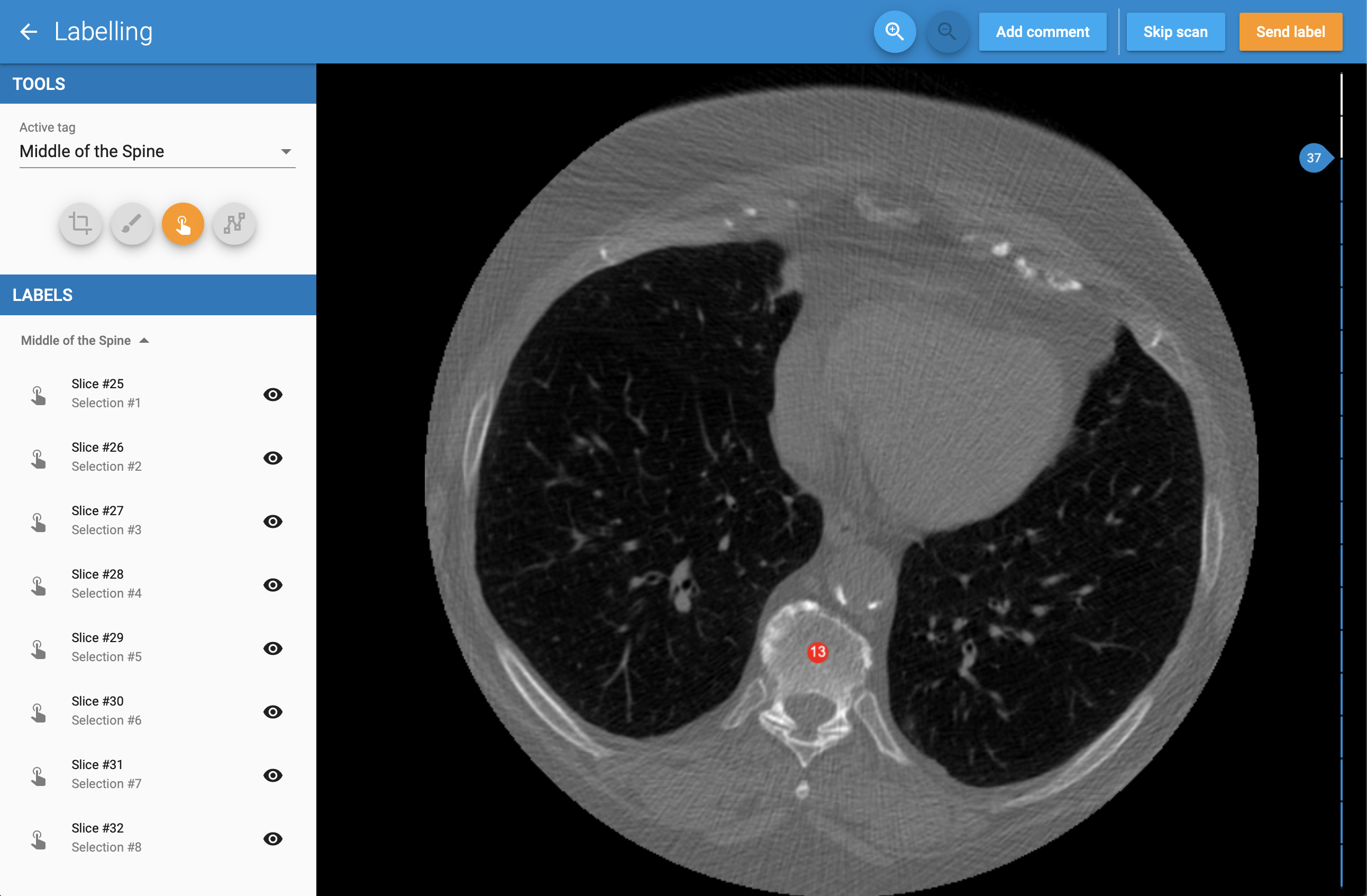
Below screenshots show how MedTagger looks like: